Day 1 :
Keynote Forum
Dr. Nabil Arrach
Progenesis Inc & the University of California, Irvine
Keynote: Reducing the Burden of Genetic Diseases via IVF and Preimplantation Genetic Diagnosis
Time : 09:30-10:15

Biography:
Dr. Nabil Arrach has 20 years of research experience in molecular genetics, both in preclinical and clinical settings. He has worked at several prestigious research centers, including the University of California, Berkeley, Sanford-Burnham Medical Research Institute and the University of California, Irvine.
Arrach was the first scientist to optimize and validate next generation sequencing for PGS and PGD in 2013. He continues to focus on technology innovation to understand and solve clinical challenges in IVF. Dr. Arrach holds a Ph.D. in Molecular and Cellular Biology.
Dr. Arrach was speaker in numerous scientific conferences including, AAB – College of Reproductive Biology (CRB), The European Society of Human Reproduction and Embryology, and The South West Embryology Summit.
Dr. Arrach is the founder of Progenesis, a leader in next generation sequencing technology for preimplantation genetic screening and diagnosis
Abstract:
It has been estimated that 5.3% of newborns worldwide will develop a genetic disorder. Certain inherited diseases seem to cluster in particular ethnic groups where consanguinity is common. For example, Canavan, Gaucher, and Maple syrup urine diseases are more common in the Ashkenazi population, while individuals of Middle Eastern descent have higher incidence of beta-thalassemia and sickle cell anemia.
Living with a genetic condition comes at a considerable financial cost to patients and dramatically impacts the healthcare system. Cystic fibrosis, for instance, is a life-threatening disease that is estimated to cost patients over $300,000 in medical expenses during their lifetime. Mitochondrial complex I deficiency is an example of a hereditary disease that has no promising treatments and is typically fatal in early childhood. Mitochondrial replacement through three-parent IVF has recently been used to minimize chance of passing mitochondrial disease to the offspring. Although this technique has yet to be legally approved in most countries, a breakthrough genome-editing technology has the potential to cure mitochondria disease. Progenesis and collaborators are currently exploring CRISPR technology to correct mitochondrial mutations in humans.
Standard practice for preventing genetic disorders in IVF involves parental carrier screening to identify disease-causing mutations, followed by preimplantation genetic diagnosis (PGD) to test embryos for the the mutation before implantation. Industry standards for carrier screening typically include a few hundred genes linked to Mendelian disorders. PGD is used after carrier screening to test embryos for one specific genetic condition, but has the potential to screen for hundreds of human diseases simultaneously. The future of inherited disease control with IVF may eliminate the need for carrier screening by replacing it with comprehensive PGD testing. These tests can significantly reduce the risks of inheriting a genetic disease, alleviate the economic burden on patients and healthcare system, and improve overall quality of life.
Keynote Forum
Dr. Manjeet Mehta
GENETIC WORLD, Molecular Genetics Laboratory, India
Keynote: Advances in our understanding of Human Genome, and how it has changed the way we diagnose, prevent & treat disease.
Time : 10:15-11:00

Biography:
Manjeet S Mehta is renowned Medical Geneticist from India par excellence. She has a rich experience of more than 3 decades in Human Genetics. She has worked with top hospitals in Bombay including Kokilaben Hospital, Jaslok Hospital, Lilavati Hospital, etc. She has set up the Genetic Department at the country’s leading chain of referral labs. She has worked with Indian Council for Medical Research (ICMR), India’s pioneer institute for biomedical research. After completing PhD, she has also gained advanced training at North York General Hospital, Toronto, and St. George’s Hospital, London. She is in the Review Board for several journals. She is an invited speaker at several conferences worldwide including the World Congress of Perinatal Medicine to be held in Belgrade this year
Abstract:
Genetics is the basic science for biology and medicine. This truth is getting wider acceptance by the medical community as nowadays any disease of any system, monogenic to multifactorial and infectious disease to cancers, needs a molecular diagnosis. There is extensive use of cytogenetic and molecular genetic techniques in cancer diagnostics, prognostication and treatment. The importance of genetic testing is accepted by clinicians as has been well proved. There is a great need of better understanding of the genetic aspects of birth defects and principles of genetic techniques so that genetic testing can be appropriately used for the benefit of evaluation of fetal anomalies and providing genetic counseling and prenatal diagnosis to the families. Genomic techniques which can analyze the whole genome in one go have made genetic testing easier and the techniques of microarray and exome/genome sequencing are being applied in clinical situations more and more frequently. It has become practically the first-tier test for most of the genetic disorders by exome sequencing for neurogenetic disorders. The cost of this latest technological marvel is within the reach of many families and the costs are likely to come down further. There are some interesting issues about next generation sequencing in medicine. Also, there is importance of knowing the significance of each nucleotide in the genome, so that the exome sequencing data can be analyzed in a more meaningful manner and with more confidence. As more and more exomes are sequenced, more and more data about pathogenic and polymorphic sequence variations is getting accumulated and these comprehensive databases will ease the challenging task of genome/exome analysis to some extent. The whole objective of diagnosis is finding a path towards curative treatment. So, at this juncture, when the clinicians are gradually getting prepared for molecular medicine, the scientific community is putting together some more pieces of the extremely complex puzzle of human physiology and pathology using genomic techniques. Medicine will take some big leaps in the next decade or two
- Human Genetics | Genetic Diseases | Evolutionary genetics | Molecular Biology | Gene Mutation | Molecular Therapies | Bioinformatics
Session Introduction
Korbinian Grote
Genomatix AG,Germany
Title: Combining DNase Hypersensitivity data and transcription factor expression to identify putative regulatory regions in leukemia
Biography:
Korbinian Grote is a Senior Scientist at Genomatix AG. His expertise is in bioinformatics of gene regulation, neural networks and deep learning. For BLUEPRINT he coordinated the project activities at Genomatix which included the development of a processing pipeline for various omics data and the creation of a visual interface to allow non-bioinformaticians easy access to the data in a biological context.
Abstract:
Acute myeloid leukemia (AML) is the most common acute leukemia in adults, with about 2 – 2.5 million new cases per year worldwide. Only 27% of patients survive 5 years or more after being diagnosed with the disease. [1]
Within the EU-funded BLUEPRINT project (http://www.blueprint-epigenome.eu) several epigenetic markers as well as expression data were generated from blood and bone marrow samples of several AML patients.
We have used DNase-Hypersensitivity and expression data from the project to identify putative transcriptional regulators and regulatory regions by combining omics data with literature. The approach included identifying DNase-HS regions that were common among AML patients, but absent in samples from healthy individuals. We then used RNA-Seq data from the project to identify transcription factor binding sites that were differentially regulated between the case and control group. Finally we used Genomatix' MatInspector software to pick regions that had both differences in DNase-HS as well as putative binding sites for the differentially expressed transcription factors. Potential target genes were selected by a next-neighbour approach and the findings were evaluated based on gene disease associations from Genomatix' literature database
Bradford D Wilson
Howard University,USA
Title: Clinical Pharmacogenetics of CYP3A4 in MAT for OUD: a case for diversity

Biography:
Bradford Wilson is a geneticist with over a decade of experience in DNA sequencing, bioinformatics, and genomics research. Dr. Wilson has conducted research in breast and prostate cancer, hypertension, and pharmacogenomics at the National Human Genome Center and the W. Montague Cobb Laboratory at Howard University. His approach to identifying the genetics underlying the biology of health disparities leverages genetic diversity and utilizes sequence variation to elucidate the pathophysiology
Abstract:
Opioid use disorder (OUD) and the associated increase in overdose deaths has become a U.S. national health priority. Medication-assisted therapy (MAT) is a treatment approach commonly used in OUD management. Buprenorphine, a partial mu-opioid receptor agonist and kappa-opioid receptor antagonist, has been shown to be an effective MAT option for OUD management. Buprenorphine is primarily metabolized by the cytochrome P450 3A4 (CYP3A4) enzyme. The CYP3A4*1B allele confers an ultrarapid metabolizer phenotype and is found at a significantly higher frequency in African populations compared to non-African populations. This genetic difference in CYP3A4 metabolism leaves some patients being managed on buprenorphine undertreated and at an increased risk for relapse. Clinical pharmacogenomics (CPGx) testing of CYP3A4 has been shown to improve MAT outcomes in African American patients. PGx-guided buprenorphine dosing reduced the number of relapses on OUD patients exhibiting the CYP3A4*1B genotype. Reduced relapse translates into additional downstream benefits including reduction in risk of Hepatitis C and/or HIV infection. The functional significance and clinical utility of this variant demonstrate the need for diversity in CPGx studies and pharmacogenomics testing algorithms.
Recent Publications:
- Opioid Metabolizing Enzyme Allele Frequencies and Drug use in a Cohort of African American Young Adults. Bradford D. Wilson, Earl B. Ettienne, Victor Apprey, Adaku Ofeogbu, Muneer Abbas, Georgia M. Dunston, Forough Saadatmand ARC Journal of Addiction 2017 2: 4-9
- Public choice theory and rhetoric: Advancing pharmacogenomics through health policy in Africa. Earl Ettienne, LP.D., MBA, RPh, Adaku Ofoegbu, PharmD, Mary Maneno, PhD, La’Marcus Wingate, PhD, Georgia Dunston, PhD, Philip Kurian, PhD, Bradford D. Wilson, PhD, KevinNguyen, Ginikannwa Ezeude, Jeronimo Augusto, MHSA African Journal of Rhetoric 2017 9: 119-142
- Pharmacogenomics-guided policy in opioid use disorder (OUD) management: An ethnically-diverse case-based approach. Earl B. Ettienne, Edwin Chapman, Mary Maneno, Adaku Ofoegbu, Bradford Wilson, Beverlyn Settles-Reaves, Melissa Clarke, Georgia Dunston, Kevin Rosenblatt. Addictive Behaviors Reports 2017 Dec; 6: 8–14
- Next generation sequencing reveals high prevalence of BRCA1 and BRCA2 variants of unknown significance in early-onset breast cancer in African American women. Luisel J Ricks-Santi, PhD; John Tyson McDonald, PhD; Bert Gold, PhD; Michael Dean, PhD; Nicole Thompson, MS; Muneer Abbas, PhD; Bradford Wilson, PhD; Yasmine Kanaan, PhD; Georgia Dunston, PhD. Ethnicity and Disease. 2017 Apr 20:27(2):169-178
- Genetic polymorphisms in the serotonin receptor 7 (HTR7) gene are associated with cortisol levels in African American young adults. G Swanson, S Miller, A Alyahyawi, B Wilson, F Saadatmand, C Lee, G Dunston, M Abbas. F1000 Research 6:19 · January 2017
Mouza Mohammed AlFashti AlAleeli
Sheikh Khalifa General Hospital , UAE
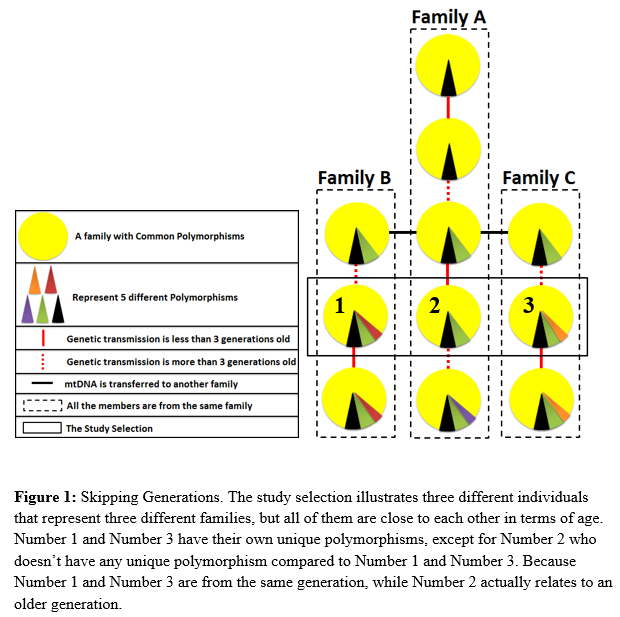
Title: The Discovery of the “Skipping Generations†Phenomenon

Biography:
Mouza Mohammed AlFashti AlAleeli received her Bachelor's degree in Medical Laboratory Science with Distinction with Highest Honor from Sharjah Higher Colleges of Technology, UAE, in August 2015. She worked for the ministry of presidential affairs as a medical laboratory scientist at Sheikh Khalifa General Hospital in Ajman, UAE, since December 2015. She is currently a member of the infection prevention and control committee of Sheikh Khalifa Medical City-Ajman and a member of the laboratory quality team of Sheikh Khalifa Medical City-Ajman. She had an experience of five months working on Mitochondrial DNA Testing in the police forensic laboratory in Sharjah, UAE. Her current research “The Significance of Applying Mitochondrial DNA Testing in Degraded Biological Evidence For The Identification of UAE Nationals to Aid in Forensic Analysis” leads her to discover the “Skipping Generations” phenomenon
Abstract:
Statement of the Problem: The demanding need to discover someone’s identity is not possible with the nuclear DNA especially when the traces are highly degraded, since the nuclear DNA is destroyed in these conditions. Only the mitochondrial DNA that is inherited maternally can survive in these compromised conditions. The purpose of this study is to find a genetic commonality between UAE nationals.
Methodology & Theoretical Orientation: 150 buccal swabs of unrelated UAE female students (approved by the UAE ID) of Sharjah Higher Colleges of Technology were collected and kept at room temperature for a period of three months or longer; to destroy the nuclear DNA, so only the mtDNA is present. mtDNA testing was performed on these buccal swabs, and it’s consisting of DNA Extraction, Real-Time quantitative PCR, Cycle sequencing and Capillary electrophoresis. The ABI PRISM®310 Genetic Analyzer capillary autosequencer [ABI PRISM® SeqScape® Soft-ware Version 2.6] was used to generate the mitochondrial DNA profiles.
Findings: From these haplotype data, a total of 229 polymorphisms were observed carefully. 106 different polymorphisms were identified out of them, and classified into unique and common polymorphisms. Interestingly, two individuals from the study subjects lacked unique polymorphisms.
Conclusion & Significance: It’s impossible for anyone to preserve their mtDNA from their great ancestors till now. The discovery of the remains of the Romanov family back in 1991concluded that the comparison of mtDNA that is more than three generations old is more likely to get at least one mutation in the current generation. Therefore, if a vertical study is done on those two individuals with their older generations, definitely they will have unique polymorphisms compared to their older generations. Those two individuals are the effect of “Skipping Generations” phenomenon, the term that I have invented to solve the mystery of having two individuals with no unique polymorphisms
Image:
Ahmed Al-Amri
National Genetic Center Royal hospital, Oman
Title: A novel LHFPL5 mutation causes autosomal recessive hearing loss in an Omani family.

Biography:
Abstract:
Hearing loss is a debilitating disorder that impairs language acquisition, resulting in disability in children and potential isolation in adulthood. Its onset can have a genetic basis, though environmental factors which are often preventable can also cause the condition. The genetic forms are highly heterogeneous and early detection is necessary to arrange appropriate patient support. Here we report the molecular basis of hereditary hearing loss in a consanguineous family with multiple affected members from Oman. Combining homozygosity mapping with whole exome sequencing identified a novel homozygous nucleotide substitution c.575T>C in the lipoma HMGIC fusion partner-like 5 gene (LHFPL5), that converted the 192nd amino acid residue in the protein from a leucine to a proline, p.(Leu192Pro). Sanger sequencing confirmed segregation with the disease phenotype as expected for a recessive condition and the variant was absent in 60,706 subjects from various disease-specific and population genetic studies as well as 50 unrelated individuals of Omani ethnicity. This study, which describes a novel LHFPL5 mutation in a family of Omani origin with hereditary hearing loss, supports previous clinical descriptions of the condition and contributes to the genetic spectrum of mutations in this form of deafness. This is the first report of a family from the Arabian Peninsula with this form of deafness
Sayyed Morteza Hosseini
Advanced Center for Biotechnology, UAE
Title: Current status and perspectives of stem cell and cloning technologies in regenerative medicine and disease therapeutics

Biography:
Sayyed Morteza Hosseini (DVM, PhD) is associated professor and senior researcher at Department of Embryology, Advanced Center for Biotechnology, Dubai, UAE. His studies have focused on the molecular and cellular aspects of reproductive biotechnology, epigenetic reprogramming and developmental reprogramming. he has authored more than 70 publications and has been involved in different projects on animal cloning and transgenesis for production of pharmaceutical drugs and also transplantable organs
Abstract:
Without any doubt, regenerative medicine is the major innovation in health care because of its great capacity to repair or replacement of damaged/diseased human cells, tissues or organs to restore normal function. Despite past limited success in the clinical translation of several promising preclinical results, the spectacular recent progress in the field of stem cells and somatic cell nuclear transfer (SCNT) technologies has laid the promising foundation for cell based therapies of disease which cannot be cured by conventional medicines.
The rapid advancement of stem cells technology can be illustrated by the progression of human embryonic stem cells (hESCs), induced pluripotent stem cells (iPS) and the use of pluripotent vs. somatic and of allogenic vs. autologous stem cells for a series of cell based therapies which have received Food and Drug Administration (FDA) approval and are now commercially available. On the other hand, the rapid advancement of SCNT technology can be illustrated by technical improvements, development of human and monkey cloned embryos and ESCs thereof as well as disease modeling and production of new lines of genetically modified cloned animals for xenotransplantation. Here, these advances and other regenerative medicine approaches currently being studied in preclinical and clinical settings will be reviewed. The popular view and the ethical issues will be addressed and finally, the perspectives, challenges and directions for the future of regenerative will be discussed.
Recent Publications :
- Lee SE, Hyun H, Park MR, Choi Y, Son YJ, Park YG, Jeong SG, Shin MY, Ha HJ, Hong HS, Choi MK, Im GS, Park EW, Kim YH, Park C, Kim EY, Park SP (2017) Production of transgenic pig as an Alzheimer's disease model using a multi-cistronic vector system. PLoS One 6;12(6).
- Liu Z, Cai Y, Wang Y, Nie Y, Zhang C, Xu Y, Zhang X, Lu Y, Wang Z, Poo M, Sun Q (2018) Cloning of Macaque Monkeys by Somatic Cell Nuclear Transfer. Cell [Epub ahead of print].
- Yao X, Liu Z, Wang X, Wang Y, Nie YH, Lai L, Sun R, Shi L, Sun Q, Yang H (2018) Generation of knock-in cynomolgus monkey via CRISPR/Cas9 editing. Cell Res [Epub ahead of print].
- Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee HS, Sritanaudomchai H, Masterson K, Larson J, Eaton D, Sadler-Fredd K, Battaglia D, Lee D, Wu D, Jensen J, Patton P, Gokhale S, Stouffer RL, Wolf D, Mitalipov S (2013) Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 153:1228-38.
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663-76.
- Maoa AS, Mooneya DJ (2015) Regenerative medicine: Current therapies and future directions. PNAS 112: 14452-14459.
Avni B. Santani
Director in the Division of Genomic Diagnostics at Children's Hospital of Philadelphia
Title: CLINICAL EXOME SEQUENCING IN PEDIATRICS: CHALLENGES AND OPPORTUNITIES

Biography:
Avni Santani, PhD, is the scientific director and has over seven years of experience in molecular diagnostic testing. She obtained her PhD at Texas A&M University and her clinical laboratory training in genetics at The Children’s hospital of Philadelphia. Dr. Santani holds specialty board certifications in Clinical Molecular Genetics and Clinical Cytogenetics. Her primary focus is on new test development and adoption of next generation sequencing technologies for diagnostic testing. Dr. Santani has experience in developing diagnostic tests for over 100 disease genes implicated in disorders such as hearing loss, epilepsy, Noonan syndrome, Rett syndrome, paraganglioma, neuroblastoma and others. Her training and experience make her an expert in a wide variety of genetic disorders, molecular biology applications and genomic
Abstract:
With the explosion of genomic information and novel gene discoveries, clinical laboratories are faced with critical challenges in facilitating sensitive data interpretation. In particular, laboratories are faced with developing strategies that can incorporate data in the interpretation of genetic variants and review variants over time.
Dr. Santani will share insights in the integration of NGS data in clinical diagnostics, using exome sequencing as an example. She will address the development of a comprehensive interpretation pipeline for exome sequencing, discuss challenges with data interpretation, and present case studies from the exome sequencing program at Children’s Hospital of Philadelphia
Ghada Al-Kafaji
Arabian Gulf Univeristy, Bahrain
Title: Defects of mitochondrial genome in diabetic nephropathy: Role and clinical relevance

Biography:
Dr. Ghada Al-Kafaji is an Associate Professor of Molecular Genetics in the Department of Molecular Medicine and the Director of Personalized Medicine Master Program at the College of Medicine, Arabian Gulf University, Bahrain. She obtained her PhD degree in Molecular Genetics from King's College London, University of London, UK. Following her PhD, she worked in the UK as a Postdoctoral Research Fellow at the School of Medicine, King's College London, and as an Assistant Professor of Genetics at the College of Science, University College Kensington. Currently, she is involved in lecturing and tutoring undergraduate and graduate students and supervising graduate theses. She has abundant publications in the area of molecular genetics that have been cited over 150 times. She participated as an active member in many International Scientific Associations. She acted as a potential reviewer for many journals and received several certificates of excellence in reviewing scientific articles. She also received a number of awards for best presentations and outstanding work in regional and international conferences.
Abstract:
Mitochondria play important roles in cellular energy metabolism and reactive oxygen species (ROS) generation. Hyperglycemia-induced overproduction of mitochondrial ROS contributes to mitochondrial dysfunction and the development diabetic complication, including diabetic nephropathy (DN). We investigated changes in the mitochondrial DNA copy number (mtDNA-CN), gene expression of mtDNA-encoded subunits of electron transport chain (ETC) complexes and mitochondrial biogenesis in DN.
ROS production, mitochondrial function, mtDNA-CN, gene expression of mtDNA-encoded ETC subunits and mitochondrial biogenesis regulatory factors were analysed in human mesangial cells cultured for 1, 4, and 7 days in normal and high glucose in the presence and absence of manganese superoxide dismutase mimic (MnTBAP) or catalase. Additionally, mtDNA-CN was analysed in peripheral blood of type 2 diabetes (T2D) patients with normoalbuminuria, DN patients with microalbuminuria or macroalbuminuria and healthy control subjects.
In the renal cells, high glucose induced a significant increase in ROS production, which was accompanied by a progressive decrease in ATP. mtDNA-CN, expression of mtDNA-encoded genes and mitochondrial biogenesis were increased at 1 day in high glucose but were decreased at 4 and 7 days. Treatment of cells with MnTBAP or catalase during high-glucose incubation attenuated ROS production and all these changes. In the subject groups, peripheral blood mtDNA-CN was significantly lower in DN patients compared with T2D patients and controls, declined with the severity of DN, and showed a significant diagnostic ability to differentiate DN patients from T2D patients and healthy controls. Lower mtDNA-CN was independently associated with the progression of DN, negatively correlated with albuminuria and conventional risk factors of DN, and positively correlated with eGFR.
Our data show that defects of mitochondrial genome paly important role in DN. Protection of mitochondria from high glucose-induced ROS may provide a potential approach to retard the development of DN. Our data also propose the mtDNA-CN as a novel blood biomarker for the early diagnosis of DN and the significance of decreased mtDNA-CN as another risk factor in the development of DN.
Recent publications
- Al-Kafaji G, AlJadaan A, Kamal A, Bakhiet M (2018). Peripheral blood mitochondrial DNA copy number is a potential new biomarker for diabetic nephropathy in type 2 diabetes patients. International Journal of Molecular Medicine. In press, Manuscript number: 204218.
- Al-Kafaji G, Sabry MA, Skrypnyk C (2016). Time-course effect of high glucose-induced reactive oxygen species on mitochondrial biogenesis and function in human renal mesangial cells. Cell Biology International 40(1):36-48.
- Al-Kafaji G, Sabry MA, Bakhiet M (2016). Increased expression of mitochondrial DNA-encoded genes in human renal mesangial cells in response to high glucose-induced reactive oxygen species. Molecular Medicine Reports 13(2):1774-80.
- Al-Kafaji G, Golbahar J (2013). High glucose-induced oxidative stress increases the copy number of mitochondrial DNA in human mesangial cells. Biomedical Research International 2013:754946.
