Day 2 :
Keynote Forum
Dr. Nabil Arrach
Progenesis Inc & the University of California, Irvine
Keynote: Reducing the Burden of Genetic Diseases via IVF and Preimplantation Genetic Diagnosis

Biography:
Dr. Nabil Arrach has 20 years of research experience in molecular genetics, both in preclinical and clinical settings. He has worked at several prestigious research centers, including the University of California, Berkeley, Sanford-Burnham Medical Research Institute and the University of California, Irvine.
Arrach was the first scientist to optimize and validate next generation sequencing for PGS and PGD in 2013. He continues to focus on technology innovation to understand and solve clinical challenges in IVF. Dr. Arrach holds a Ph.D. in Molecular and Cellular Biology.
Dr. Arrach was speaker in numerous scientific conferences including, AAB – College of Reproductive Biology (CRB), The European Society of Human Reproduction and Embryology, and The South West Embryology Summit.
Dr. Arrach is the founder of Progenesis, a leader in next generation sequencing technology for preimplantation genetic screening and diagnosis
Abstract:
It has been estimated that 5.3% of newborns worldwide will develop a genetic disorder. Certain inherited diseases seem to cluster in particular ethnic groups where consanguinity is common. For example, Canavan, Gaucher, and Maple syrup urine diseases are more common in the Ashkenazi population, while individuals of Middle Eastern descent have higher incidence of beta-thalassemia and sickle cell anemia.
Living with a genetic condition comes at a considerable financial cost to patients and dramatically impacts the healthcare system. Cystic fibrosis, for instance, is a life-threatening disease that is estimated to cost patients over $300,000 in medical expenses during their lifetime. Mitochondrial complex I deficiency is an example of a hereditary disease that has no promising treatments and is typically fatal in early childhood. Mitochondrial replacement through three-parent IVF has recently been used to minimize chance of passing mitochondrial disease to the offspring. Although this technique has yet to be legally approved in most countries, a breakthrough genome-editing technology has the potential to cure mitochondria disease. Progenesis and collaborators are currently exploring CRISPR technology to correct mitochondrial mutations in humans.
Standard practice for preventing genetic disorders in IVF involves parental carrier screening to identify disease-causing mutations, followed by preimplantation genetic diagnosis (PGD) to test embryos for the the mutation before implantation. Industry standards for carrier screening typically include a few hundred genes linked to Mendelian disorders. PGD is used after carrier screening to test embryos for one specific genetic condition, but has the potential to screen for hundreds of human diseases simultaneously. The future of inherited disease control with IVF may eliminate the need for carrier screening by replacing it with comprehensive PGD testing. These tests can significantly reduce the risks of inheriting a genetic disease, alleviate the economic burden on patients and healthcare system, and improve overall quality of life
- Molecular Modeling | Gene Sequencing | Pharmacogenetics | Immunogenetics | Translational Medicine | Epigenetics | Stem cell Transplantation
Session Introduction
Marlena Szalata
Poznan University of Life Sciences, Poland
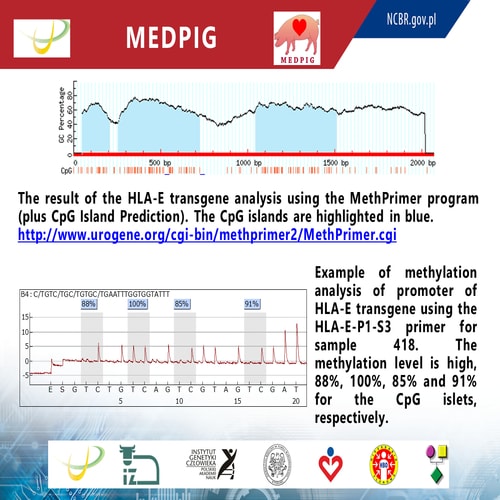
Title: Methylation level of HLA-E transgene in the xenogeneic tissues of transgenic pigs

Biography:
Marlena Szalata works in biotechnology field with the main emphasis on transgenesis of animals and plants. She is involved in purification and characterization of recombinant proteins obtained in prokaryotic and eukaryotic systems like plants and animal bioreactors. Marlena Szalata has expertise in molecular diagnostics of human, animal and plants genes. Recently she is involved in analysis of methylation level of selected human, animal and plant genes.
Abstract:
Statement of the Problem:
Epigenetics involves studies of changes in gene expression not associated with changes at DNA sequence level. The best characterized epigenetic mechanism involved in control of gene expression is DNA methylation. Methylation of cytosines in CpG island within promoters usually causes gene silencing. In transgenic animals we are interested not only in introduction of transgene but also in its activity. We have prepared transgenic pigs for xenotransplantation purposes with decreased recognition by human immune system by introduction of the human HLA-E gene under elongation factor 1 alpha promoter (EF-1α). Activity of HLA-E transgene was already confirmed on molecular level and flow cytometry. The purpose of this study was evaluation of specific methylation of HLA-E transgene.
Methodology & Theoretical Orientation: Genomic DNA was modified by bisulfite conversion and PCR products were analysed using pyrosequencing method. The most important steps were DNA isolation and DNA bisulfite conversion. Pyrosequencing enabled quantification of the specific methylation level of transgene.
Findings: The total level of methylation of the HLA-E transgene promoter was approximately 90% (range 67-100%) and the HLA-E gene 73.79% (29-100%). The obtained results of the HLA-E transgene methylation were compared to the results of transgene expression, which showed that the level of transgene expression in transgenic animals was stable, regardless of pig generation. This suggest, that level of methylation of transgene promoter does not affect directly its biological activity estimated by flow cytometry.
Financed by National Centre for Research and Development (no INNOMED/I/17/NCBR/2014) within framework of INNOMED programme Development of an innovative technology using transgenic porcine tissues for biomedical purposes. Acronym MEDPIG.
Image:
Recent Publications (minimum 5)
- Cheung NKM, Nakamura R, Uno A, Kumagai M, Fukushima HS, Morishita S, Takeda H (2017) Unlinking the methylome pattern from nucleotide sequence, revealed by large-scale in vivo genome engineering and methylome editing in medaka fish. PLoS Genet 13(12):e1007123.
- Gödecke N, Zha L, Spencer S, Behme S, Riemer P, Rehli M, Hauser H, Wirth D (2017) Controlled re-activation of epigenetically silenced Tet promoter-driven transgene expression by targeted demethylation. Nucleic Acids Research 45(16):e147.
- Hryhorowicz M, Zeyland J, Słomski R, Lipiński D (2017) Genetically Modified Pigs as Organ Donors for Xenotransplantation. Molecular Biotechnology 59(9-10):435-44
- Kimsa-Dudek M, Strzalka-Mrozik B, Kimsa MW, Blecharz I, Gola J, Skowronek B, Janiszewski A, Lipinski D, Zeyland J, Szalata M, Slomski R, Mazurek U (2015) Screening pigs for xenotransplantation: expression of porcine endogenous retroviruses in transgenic pig skin. Transgenic Research 24(3):529-536.
- Nuo MT, Yuan JL, Yang WL, Gao XY, He N, Liang H, Cang M, Liu DJ (2016) Promoter methylation and histone modifications affect the expression of the exogenous DsRed gene in transgenic goats. Genetics and Molecular Research 15(3).
Ryszard Slomski
Poznan University of Life Sciences, Poland
Title: Development of an innovative technology using transgenic porcine tissues for biomedical purposes

Biography:
Professor Ryszard Slomski, is a head of the Department of Biochemistry and Biotechnology at Poznan University of Life Sciences and Professor at Institute of Human Genetics of the Polish Academy of Sciences in Poznan. His main research focus on: 1) preparation of gene constructs containing human genes for expression in animal cells, including large animals and preparation of other gene constructs in prokaryotic and eukaryotic systems; 2) characteristics of novel human, animal and plant genes, 3) paternity and relationship testing based on DNA studies; 4) molecular diagnostics of Duchenne muscular dystrophy (DMD), familial polyposis of the colon (FAP), thyroid cancer and Crohn’s disease. Professor Slomski is involved in development of an innovative technology of biomedical applications for tissues of transgenic pigs, acronym MEDPIG and development of cannabinoid collection technology from hemp with low THC content as means of treatment of pain in cancer patients, acronym ONKOKAN
Abstract:
Statement of the Problem:
The growing shortage of available organs is a major problem in transplantology. The promising solution could be xenotransplantation, i.e., the use of cells, tissues and organs of domestic pig. However, xenogeneic transplantation from pigs to humans involves high immune incompatibility and a complex rejection process. The rapid development of genetic engineering techniques enables genome modifications in pigs that reduce the cross-species immune barrier. The purpose of this study is to develop the technology to use transgenic pigs for biomedical purposes. The Project is an alternative to studies using stem cells and work aiming at the production of artificial organs.
Methodology & Theoretical Orientation: Development of an animal breeding system and the collection of skin, valves and blood vessels from transgenic pigs. The project involves also production of polytransgenic pigs using reproduction biotechnology methods. Very important was development and characteristics of gene constructs preventing xenotransplant rejection and multiparametric characteristics of transgenic animals. Tissues of transgenic animals were used for treatment of human cardiovascular diseases using vessels and cardiac valve bioprostheses. Biological dressings were obtained from skin of transgenic pigs.
Findings: Important issues in production of transgenic animals are characteristics of transgene, transfection and cell transformation, detection of transgene integration, mapping of transgene, passing specific traits to the offspring, homozygote selection, analysis of transgene activity and function. Fully characterized transgenic animals carrying three or more modifications may be used as sources of skin, heart valves or vessels.
Conclusion & Significance: Transgenic animals were generated using traditional methods and new genome editing technologies. Tissues of transgenic animals used in experimental studies showed growing potential in use as alternative treatment of human. This work was supported by grant No. INNOMED/I/17/NCBR/2014 from the National Centre for Research and Development.
Image:
Recent Publications (minimum 5)
- Hryhorowicz M, Lipiński D, Zeyland J, Słomski R (2017) CRISPR/Cas9 Immune System as a Tool for Genome Engineering. Archivum Immunologiae et Therapiae Experimentalis 65:233-240.
- Hryhorowicz M, Zeyland J, Słomski R, Lipiński D (2017) Genetically Modified Pigs as Organ Donors for Xenotransplantation. Molecular Biotechnology 59:435-444.
- Kimsa-Dudek M, Strzalka-Mrozik B, Kimsa MW, Blecharz I, Gola J, Skowronek B, Janiszewski A, Lipinski D, Zeyland J, Szalata M, Slomski R, Mazurek U (2015) Screening pigs for xenotransplantation: expression of porcine endogenous retroviruses in transgenic pig skin. Transgenic Research 24:529-536.
- Wilczek P, Lesiak A, Niemiec-Cyganek A, Kubin B, Slomski R, Nozynski J, Wilczek G, Mzyk A, Gramatyka M (2015) Biomechanical properties of hybrid heart valve prosthesis utilizing the pigs that do not express the galactose-α-1,3-galactose (α-Gal) antigen derived tissue and tissue engineering technique. Journal of materials science. Materials in Medicine 26:5329.
- Cierpka L, Ryszka F, Dolińska B, Smorąg Z, Słomski R, Wiaderkiewicz R, Caban A, Budziński G, Oczkowicz G, Wieczorek J (2014) Biolasol: novel perfusion and preservation solution for kidneys. Transplantation Proceedings 46:2539-2541.
Namrata Londhe
Kokilaben Dhirubhai Ambani Hospital, India
Title: Determination of genetic cause in cases of developmental delay- a retrospective study in Indian cohort
Biography:
Dr. Namrata Londhe has done her doctoral work in Human Molecular Genetics at Bhabha Atomic Research centre, Mumbai. She is currently working in Kokilaben Dhirubhai Ambani Hospital and Medical Research centre as an expertise in cytogenetics and FISH based assays in constitutional, hematological and solid tumors. During her academic and industrial tenure, she has worked on several cytogenetic and molecular biology techniques and has presented her work at several national and international conferences
Abstract:
Statement of the Problem: To elucidate the genetic component in patients with developmental delay (DD) through 5 year retrospective data analysis. After the initial workup by the pediatrician, the patients were referred to the department for cytogenetic and molecular testing.
Methodology & Theoretical Orientation: Most detrimental form of developmental delay is global DD which is presented as overall absence of the required milestone in the child including mental, cognitive as well as motor delay. The advent of cytogenetic and molecular biology techniques has been helpful in understanding the genetic etiology of these conditions. DD is affecting 1:150 children. Cases with following indications like Attention Deficit Disorder, Attention Deficit Hyperactivity Disorder, Global DD, Pervasive Developmental Disorder, Pervasive Developmental Disorder-Not Otherwise Specified, Syndromic, Intellectual Disability, Learning Disability, Specific DD, speech delay etc. were all included in the data analysis.
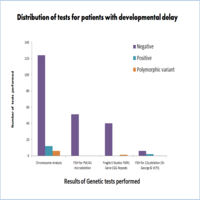
Findings: A total of 199 cases (age ranging from 5 months- 22years) were studied. Cytogenetic analysis was performed on 142 patients and chromosomal abnormalities were observed in 8.5% (12/142) of the cases. FISH assays (For Praderwilli/angelman syndrome, Digeorge, etc) were performed in 59 cases, positive results were obtained in 3.3% (2/59). Molecular analysis (Fragile X analysis, Rett syndrome, etc) was done for 44 subjects. One case 2.4% (1/44) revealed an intermediate phenotype for Fragile-X studies. Chromosomal abnormalities were found in 8.4% (12/142) cases. This observations can be further classified as structural abnormalities in 66.7% (8/12), numerical abnormalities in 25% (3/12) and a combination both abnormalities was seen in 8.3% (1/12) of the cases. Polymorphic variants were found in 4.2% (6/142) of the cases.
Conclusion & Significance: Cytogenetic and molecular tests could successfully elucidate the genetic components implicated in 7.5% of the total cases. As chromosomal analysis can detect the anomalies up to 5MB resolution, advanced techniques like Microarray studies would be more helpful in detecting the cryptic subtle rearrangements in unsolved cases
Image:
Recent Publications:
- John B. Moeschler. Comprehensive Evaluation of the Child With Intellectual Disability or Global Developmental Delays. Pediatrics 2014;134:e903–e918.
- PanelLisa G. Shaffer. Innovations in the Early Diagnosis of Chromosomal Disorders Associated with Intellectual Disability. International Review of Research in Developmental Disabilities. vol 40, 2011, Pages 211-228. https://doi.org/10.1016/B978-0-12-374478-4.00008-3
- David T. Miller, et al. Consensus Statement: Chromosomal Microarray Is a First-Tier Clinical Diagnostic Test for Individuals with Developmental Disabilities or Congenital Anomalies. Am J Hum Genet. 2010 May 14; 86(5): 749–764. doi: 10.1016/j.ajhg.2010.04.006
- Dave BJ, et al. Role of Cytogenetics and Molecular Cytogenetics in the diagnosis of genetic imbalances. Semin Pediatr Neurol. 2007 Mar;14(1):2-6.
- WS Meschino. The child with developmental delay: An approach to etiology. Paediatr Child Health 2003;8(1):16-19
Serhat Seyhan
Bakirkoy Dr. Sadi Konuk Training and Research Hospital, Turkey
Title: Correlation between Expressions of IL-3, IL-6 and IL-11 Genes and Chronic Myeloid Leukemia

Biography:
Serhat Seyhan was born in istanbul in 1987. He graduated from Cumhuriyet University faculty of Medicine in 2011. He completed Karadeniz Technical University medical genetic phd in 2016, which he started in 2012. He is currently working as a medical geneticist in Istanbul Bakirkoy Dr. Sadi Konuk Training and Research Hospital
Abstract:
Statement of the Problem:
Chronic myeloid leukemia (CML) is a hematopoietic malignancy originating from transformation of a hematopoietic stem cell. BCR-ABL gene that resulted from a translocation between chromosomes 9 and 22 is detected in the majority of CML patients. IL-3, IL-6 and IL-11 are cytokines that affects the development of myeloid cells. The aim of this study to evaluate the relationship between IL-3, IL-6 and IL-11 and CML development.
Methodology & Theoretical Orientation: This study was perfomed using peripheral blood from 33 patients followed in our hematology clinic at Karadeniz Technical University Medical School obtained with a diagnosis of CML. We get peripheral blood from 34 healthy people that examined in Karadeniz Technical University Medical Schools clinics. RNA was isolated from peripheral blood and cDNA is synthesized. Then cDNA expression analysis was performed. Findings: IL-3, IL-6 and IL-11 gene expressions significantly decreased in CML patients with high expression of BCR-ABL gene compared to healthy individuals. After major molecular response IL-3, IL-6 and IL-11 gene expressions increased compared with the initial diagnosis of CML.
Conclusion & Significance: According to the our results; increase of BCR-ABL gene expression in CML patients may provide the proliferation of neoplastic cells without effective IL-3, IL-6 and IL-11 gene expressions in normal hematopoiesis. Ä°nverse relationship in our CML patients are available between BCR-ABL expression and IL-3, IL-6 and IL-11 expressions. Studies for IL-3, IL-6 and IL-11 gene expressions are needed to determine the relationship with BCR-ABL expression level and to be used follow up of patients with CML
Recent Publications (minimum 5)
- Pehlivan M, Sahin HH, Pehlivan S, Ozdilli K, Kaynar L, Oguz FS (2014) Prognostic importance of single-nucleotide polymorphisms in IL-6, IL-10, TGF-beta1, IFN-gamma, and TNF-alpha genes in chronic phase chronic myeloid leukemia. Genetic testing and molecular biomarkers 18 (6):403-9.
- Welner RS, Amabile G, Bararia D, Czibere A, Yang H, Zhang H (2015) Treatment of chronic myelogenous leukemia by blocking cytokine alterations found in normal stem and progenitor cells. Cancer cell 27 (5):671-81.
- Apperley JF (2015) Chronic myeloid leukaemia. Lancet 385 (9976):1447-59.
- Shet AS, Jahagirdar BN, Verfaillie CM (2002) Chronic myelogenous leukemia: mechanisms underlying disease progression. Leukemia 16 (8):1402-11.
- Ren R (2002) The molecular mechanism of chronic myelogenous leukemia and its therapeutic implications: studies in a murine model. Oncogene 21 (56):8629-42.
- Melo JV, Deininger MW (2004) Biology of chronic myelogenous leukemia--signaling pathways of initiation and transformation. Hematology/oncology clinics of North America 18 (3):545-68, vii-viii.
Maya N Gaiozishvili
Ivane Javakhishvili Tbilisi State University,Georgia
Title: GST Genes (GSTM1 and GSTT1) Polymorphism in Georgian Population

Biography:
Maya Gaiozishvili is PhD in biology. She is Assistant-Professor at department of genetics, Ivane Javakhishvili Tbilisi State University. The field of her interest is the personalized medicine, human genetics, exactly, aging and medical genetics. She is investigator in grant projects, which concerns investigation of some genes polymorphism in Georgian population, also genetic parameters in different diseases. By her participation was determined the polymorphism of genes involved in warfarin metabolism, in Georgian population. Also, was determined the types of mutations and epigenetic variability - namely the degree of heterochromatinization, during the aging, atherosclerosis, cardiomyopathy, breast and lung cancers, in Georgian population. In some cases, the possibility of correction of altered genetic parameters by different agents. The results of the study are published in scientific journals.
Abstract:
The glutathione S-transferase (GST) enzyme system constitutes a family of multifunctional enzymes which play an important role in biotransformation and detoxification of many different xenobiotic. Human cytosolic GSTs are polymorphic, and have ethnic-dependent polymorphism frequencies, and have been associated with several types of diseases. In the double-null variant of GSTM1 and GSTT1 are respectively associated with a higher risk of different forms of liver injury, cancer, cardiovascular diseases etc[1-3].
Comparisons between GST null genotype frequencies in the worldwide populations show different patterns in Asian, African, and European populations.
Some detailed studies of GST variants in various geographic regions can increase the knowledge about relationship between the ethnicity and the prevalence of certain diseases[4]. Thus, it becomes necessary to consider the genotypic differences for reducing the risk of anti-tuberculosis drug-induced Liver injury[5].
The aim of our research was to determine the polymorphism of GSTM1 and GSTT1 genes in Georgian population.
In the studies was be used peripheral blood samples obtained from individuals randomly selected groups. The GSTM1 and GSTT1 null genotypes was investigated with the help of an Ese-Quant tube scanner-by SmartAmp Method[6].
The study of GST genes polymorphism in Georgian population revealed that 88% of individuals have positive genotypes of GSTT1 and GSTM1, and 12% of individuals have null genotype only by one of this gene - GSTT1 or GSTM1. Out of these, GSTT1(-) null genotype was observed in 4% of individuals and GSTM1(-) have 8% of individuals. It should also be noted that there was not observed double null genotype (GSTT1(-), GSTM1(-)) in investigated group of individuals.
In the next stage of our research we will investigate the relation between the null genotypes of GSTM1 and GSTT1 and risk of development drug-induced hepatotoxicity in TB patients from Georgianpopulation.
The results will play the most important role in personalized medicine, in the appointment and management of drugs and for prevention of adverse drug reactions in patients of Georgian population.
Recent Publications (minimum 5)
- Rafiee L., Saadat I., Saadat M. (2010) Glutathione S-transferase polymorphisms (GSTM1, GSTT1 and GSTO2) in three Iranian population. Mol Biol Rep. 37, 155-158.
- Wang J.,Zou L., Huang S., et al. (2010) Genetics polymorphism of glutation-S-transferaze genes GSTM1and GSTT1 and risk coronary heart disease. Mutagenesis. 25:365-369.
- Nafissi S., Saadat I., Saadat M. (2011) Genetics polymorphism of glutation-S-transferaze Z1 in Iranian population. Mol Biol Rep. 38:3391-3394.
- Piacentini S., Polimanti R., Porreca F. (2011) GSTT1 and GSTM1 gene polymorphisms in European and African populations. Mol Biol Rep. 38:1225–1230.
- Gupta V., Singh M., Amarapurkar D. et al. (2013) Association of GST null genotypes with anti-tuberculosis drug induced hepatotoxicity in Western Indian population. Ann Hepatol. 12(6):959-965.
- Okada R, Ishizu Y, Endo R, Lezhava A, et al. (2010) Direct and rapid genotyping of glutathione-S-transferase M1 and T1 from human blood specimens using the SmartAmp2 method. Drug Metab Dispos. Oct; 38(10):1636-1639.
Tamar J Buadze
Ivane Javakhishvili Tbilisi State University, Georgia
Title: Genome Instability in Pulmonary Tuberculosis

Biography:
Tamar Buadze is PhD in biology. She is Senior Scientist in department of genetics, Ivane Javakhishvili Tbilisi State University. The field of her interest is the human genetics, exactly, medical genetics. She is principal investigator in grant projects, which concerns investigation of genetic parameters in different diseases, such as cardiomyopathy, tuberculosis and tumor - namely breast cancer. By her participation was determined the types of mutations and epigenetic variability - namely the degree of heterochromatinization, during the aging, atherosclerosis, cardiomyopathy, breast and lung cancers, in Georgian population. In some cases, the possibility of correction of altered genetic parameters by different agents. The results of the study are published in scientific journals.
Abstract:
Pulmonary Tuberculosis (PT) is classified as a disease with a hereditary predisposition. Genetic factors contribute to the outcome of primary PT infection, with an estimated heritability of more than 50%[1,2]. The genotype of an organism plays an important role in the development of PT diseases. Numerous studies have been performed to identify genetic factors responsible for variation in PT susceptibility[3]; however, none of the candidate genes was associated with susceptibility to active PT[4,5]. Therefore, as with all the pathologies related to this group, particular importance is attached to finding those markers that enable early detection of high risk groups of the disease in the population. Data on the variability of such informative and important functional parameters, which are associated with the epigenetic processes in PT, are practically absent.
The aim of our study was to evaluate genetic and epigenetic variation of the genome in patients with sensitive Pulmonary Tuberculosis (PT) before and after treatment, under the effect of peptide bioregulator–Ala-Glu-Asp-Gly.
In lymphocyte cultures from patients with sensitive primary PT were studied: facultative heterochromatin (sister chromatid exchanges - SCE) and mutation(chromosome aberrations).
We determined: There was an epigenetic alteration of functional parameters of the genome in PT before treatment. The level of heterochromatin decreased in the telomeric regions of chromosomes - A1,A2,B,C,D,F and G (in control it was high) and increased in the middle regions of chromosomes - A1,B,C,E,F, and G (in control it was reduced); There was a high level of somatic recombination; Revealed an increase of the frequency of cells with chromosome aberrations. The bioregulator (Ala-Glu-Asp-Gly) could be used as an aid in the prevention and treatment of tuberculosis.
The results of our study - redistribution of heterochromatin from the telomeric to middle chromosome arms and increasing of chromosome aberrations make it possible to define a sensitive form of PT, and then monitor the results of treatment
